
News & Press
A search for xylan debranching enzymes



10 February 2022
EnXylaScope aims to develop tools to accelerate the discovery of xylan debranching enzymes, prepare biodiversity for enzyme screening activities, as well as search for novel enzyme candidates. The screening activities will target existing and new samples, either by sequence-based mining approaches of public or in-house data bases (as well as new datasets created within the project) or apply functional screening procedures on samples, strain isolates or cloned metagenomic libraries in searches for novel enzymes able to debranch xylan polymers.
Xylan is a very abundant lignocellulose polymer in nature, found in many plants. However, due to several reasons, xylan is yet highly underexploited, and the EnXylaScope project aims to provide tools to overcome some of the challenges and limitations existing today connected to the usage of xylan. For example, for xylan to be incorporated in several consumer products and hence replace less sustainable components and allowing a greener option for consumers, the polymer needs different modifications. These modifications can be e.g. debranching of the polymer, and efficient and selective enzymes are the route for a sustainable process to produce xylan polymers meeting the needs. By debranching xylan, the polymer gathers unique properties such as reduced water solubility and enhanced viscosity, being beneficial for incorporation in e.g. skin care products, nutraceuticals and more. However, while some xylan modifying enzymes, such as laccases and lipases for polymer grafting are readily available, suitable enzymes for xylan debranching are not yet available, acting mostly on oligomers and not polymers and/or present a very narrow operating range and thereby making them unsuitable for industrial processes. Hence, the need for novel enzymes functioning in reactions for xylan debranching is highly desirable and a key point to address in the road to increase usage of xylan as a biopolymer in consumer products.
The EnXylaScope project has highlighted four different xylan debranching enzyme classes; i) GH115-α-glucuronidase, ii) α-1,2-L-arabinofuranosidase, iii) acetyl xylan esterase and iv) ferulic esterase. The action of these enzymes on the xylan polymer and its sidechains are shown in the figure below.
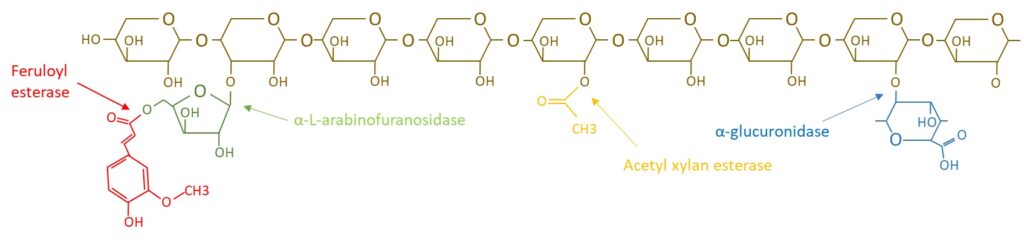
The debranching of xylan will generate a polymer with higher viscosity and lower water solubility, as well as production of the side chain products, such as e.g. ferulic acid for the activity of feruloyl esterase. However, it is the properties of the polymer that is desired as a product component, even though the production of side chain products upon enzyme-aided debranching might be of relevance for functional assay development, i.e. procedures to detect enzyme activity.
Through a dedicated work package, the EnXylaScope project aims to develop tools for discovery of xylan debranching enzymes, as well as collect samples (biodiversity) and use these in searches for new enzyme candidates to evaluate further use in xylan debranching reactions.
Novel enzyme candidates can be searched both by a functional approach (i.e. by looking for the desired functions in a biodiversity sample), or by a sequence-based approach, meaning that the sequences of the biodiversity investigated is mined for enzyme encoding genes with a certain level of similarity to known enzyme genes, assembling enzyme candidate shortlists from sequence data, to be taken further for functional evaluation. The sequence-based approach is firstly applied to available databases, both public ones as well as in-house at consortium partners. In addition, new sequence datasets are to be gathered during the project, e.g. by sequencing of DNA isolated from Svalbard soil samples as well as enriched environmental fermentation samples and potentially genome sequences of selected isolates. These will also be used in sequence mining approaches. For functional screening, one uses either an environmental sample directly (such as the enriched environmental fermentation sample described above), and evaluate if there are desired enzymatic functions therein. Alternatively, the DNA of the sample(s) is isolated and cloned as a metagenomic library (typically carried by an Escherichia coli strain) for functional screening approaches. The clone library is then replicated and used in functional screening analysis to find cloned genes with the desired function(s). Once clones producing functional enzymes are found, the genes encoding the enzymes of interest are identified, and can be transferred to efficient expression systems for enzyme production and characterisation.
For efficient and relevant functional screening of biodiversity (samples, isolates or cloned metagenomic libraries), functional screening protocols targeting the relevant enzyme functions are crucial, and in order to be able to screen large volumes of biodiversity a high throughput screening (HTS) procedure is very desirable. One of the activities EnXylaScope is to develop such HTS pipelines for accelerated discovery of xylan debranching enzymes. Enzymatic assays targeting the substrates of interest in combination with the enzyme activities (and substrate spectrum) of interest is not presently available, and development and testing of such procedures is of high relevance for success in novel enzyme discovery. Several approaches can be used to develop such assays, relying on different detection systems and pipelines, such as MS-based detection, monitoring of viscosity and/or turbidity, plate assays with xylan substrates (immune assays and spectrophotometric assays), usage of fluorometric short-chain substrates and product detection by thin layer chromatography (TLC). The targeted approaches in Enxylascope are immunoassays, spectrophotometric (plate assays) and MS-based detection.
Since the EnXylaScope project is within its first year, work on these tasks has been initiated and ais ongoing, with major results being generated at a later stage.
- Assay development is ongoing, applying different strategies
- Preparation of biodiversity for screening both by sequence-based approaches as well as functional-approaches has been ongoing since the beginning of the project. To date metagenomic libraries of >40 000 fosmid clones have been constructed, and further samples obtained from enrichment methods are in the process of being subjected to DNA isolation, DNA sequencing and library cloning.
- Both public as well as in-house databases have been subjected to sequence mining, searching for all four enzyme classes, but initially focusing on generating a candidate shortlist for α-1,2-L-arabinofuranosidase enzyme candidates. Post mining analysis involve filtration of hits, extraction of open reading frames (ORFs), multiple sequence alignment (MSA), sequence similarity networks (SSN) and BLASTing. The first candidate shortlist will be presented at month 10 of the project, being channelled into WP3 for gene expression and enzyme characterisation.
WP2 of EnXylaScope will in the nearest future focus on preparation of biodiversity for screening for new enzymes and continuation of the ongoing sequence mining activities, expanding the searches to more sequence databases as well as create shortlist for all 4 enzyme classes. In addition, high focus will be given to the establishment and, further later into the project, the execution of functional screening for relevant xylan debranching enzymes, whereafter these are transferred into WP3 for production and characterisation.








